Kojak
Free, open-source application for identification of cross-linked peptides from mass spectra.
Highlights:
- Use virtually any stable cross-linker chemistry
- Analyze isotopically labeled or non-labeled data
- Supports open data formats (including mzXML and mzML)
- Pre-compiled binaries get you started quickly
Current Release
Version 2.1.0
Released on October 16 2023
Precompiled binaries provided for Windows and Linux. Free and open source, can be compiled on virtually any operating system including Windows, Linux, and MacOS.
Get It Now » Quick Start Guide »
Read the Latest News
New Inputs and Outputs
Oct 16 2023
This new version of Kojak has some cool new features that we will continue to develop in the coming months. Among them is support for the .mzMLb format ...
Read More »
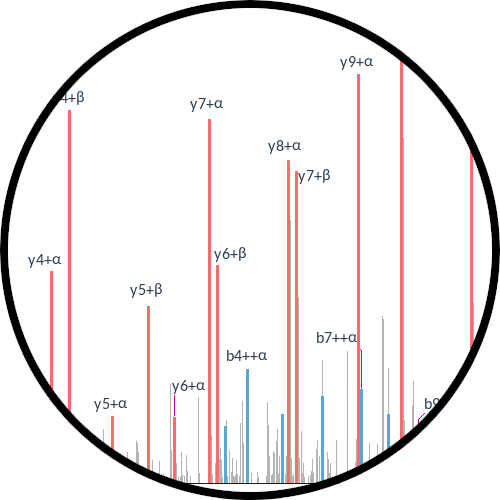
Kojak Spectrum Viewer
Visualize MS/MS spectral results from Kojak. Multi-platform graphical user interface and publication quality exporter of cross-linked spectral images.

Trans-Proteomic Pipeline
Multi-platform graphical interface for using Kojak, combined with downstream validation tools to estimate probabilities and FDR of Kojak results.

ProXL: Protein XL Database
Analyze, visualize, and share your protein cross-linking data. Import Kojak results, combine multiple analyses, mine protein links, and produce 3D protein structure overlays.
